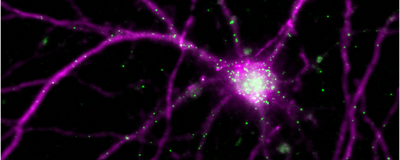
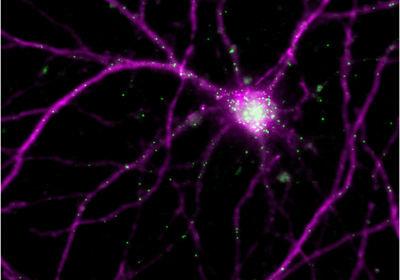
ABOVE: Arc mRNAs (green) are synthesized in the neuron’s nucleus and localized to the cell’s dendrites. Sulagna Das
For a memory to be consolidated long term, specific synapses connecting neurons must be strengthened, which requires the presence of certain proteins. Yet, some of these proteins and their mRNAs have a short half-life. Using high temporal and spatial resolution imaging, researchers have resolved this paradox for one of these memory associated molecules. Their study, published recently in Neuron, reports that a single stimulus in mouse cells and brain tissue triggers a series of transcription and translation cycles over time, promoting the formation of long-lasting memories.1
“It's been a long-held dogma in the field that protein synthesis is a necessary molecular substrate for memory consolidation,” said Stony Brook University neuroscientist Prerana Shrestha, who did not participate in this study.
Until recently, researchers “lacked the tools to study spatial-temporally resolved protein synthesis happening in real time in subcellular compartments. [This work] provides a beautiful example of what we can do with revolutionary high-resolution imaging tools,” she added.
Among the genes essential for synaptic remodeling during memory formation is the activity-regulated cytoskeletal associated (Arc) gene, whose mRNAs and proteins have a half-life of about an hour. Reconciling how these molecules produce changes on scales of hours or even days was one of the primary motivations for this study, said Sulagna Das, an RNA biologist and neuroscientist at the Albert Einstein College of Medicine and coauthor of this work.
See also “Asthma Drug Helps Mice Retrieve Memories “Lost” to Sleep Deprivation”
Das’s team tackled the question by visualizing all of the steps, tracking the mRNA from birth, following where they go and where they localize, watching when they are translated into proteins, and determining how long the proteins stay in the synapses. For this, the team fluorescently tagged the Arc gene and followed the journey of its mRNAs and proteins for several hours within individual brain cells after one initial chemical or optogenetic stimulus, both in cultured mouse hippocampal neurons and in mouse brain tissue.
By closely spying on these molecules, Das and her colleagues found that a single initial stimulus resulted in a series of cycles of transcription. The first one was initiated by the stimulus, but the subsequent ones were induced by the translated Arc proteins themselves, suggesting the existence of a positive feedback loop.
mRNAs produced during the second transcription cycle move to the dendritic regions populated by the translated proteins of the first cycle, leading to local translation and the formation of protein hubs. “That's how you can build up a lot of protein at a single location,” Das explained, and it’s important because this mechanism gives specificity to which synapses are being stabilized for long-term information storage.
See also “Inside the Brains of Aging Dogs”
Although it remains to be tested, it is very likely that this mechanism also occurs in humans, said Shrestha. For instance, Arc is expressed in humans and it is implicated in neurological disorders, such as Alzheimer’s disease and Angelman Syndrome, which involves motor dysfunction and severe intellectual disability.2,3 There is still a long way to go for this to translate into therapeutic interventions for such diseases, she cautioned, yet for now, these findings offer “a tremendous amount of understanding of how memory consolidation works and the underlying molecular substrate.”
References
- Das S, Lituma PJ, Castillo PE, Singer RH. Maintenance of a short-lived protein required for long-term memory involves cycles of transcription and local translation. Neuron. Published online 2023. doi:10.1016/j.neuron.2023.04.005
- Wu J, Petralia RS, Kurushima H, et al. ARC/arg3.1 regulates an endosomal pathway essential for activity-dependent β-amyloid generation. Cell. 2011;147(3):615-628. doi:10.1016/j.cell.2011.09.036
- Greer PL, Hanayama R, Bloodgood BL, et al. The Angelman syndrome protein Ube3A regulates synapse development by Ubiquitinating Arc. Cell. 2010;140(5):704-716. doi:10.1016/j.cell.2010.01.026